Tutorial¶
Example in 2D Uniform Distribution¶
This example generates a 2D random uniform distribution, and then uses GriSPy to search neighbors within a given radius and/or the n-nearest neighbors
Import GriSPy and others packages¶
[1]:
import numpy as np
import matplotlib.pyplot as plt
from grispy import GriSPy
[2]:
%matplotlib inline
Create random points and centres¶
[3]:
Npoints = 10 ** 3
Ncentres = 2
dim = 2
Lbox = 100.0
np.random.seed(0)
data = np.random.uniform(0, Lbox, size=(Npoints, dim))
centres = np.random.uniform(0, Lbox, size=(Ncentres, dim))
Build the grid with the data¶
[4]:
gsp = GriSPy(data)
Set periodicity. Periodic conditions on x-axis (or axis=0) and y-axis (or axis=1)
[5]:
periodic = {0: (0, Lbox), 1: (0, Lbox)}
gsp.set_periodicity(periodic)
[5]:
GriSPy(N_cells=20, periodic={0: (0, 100.0), 1: (0, 100.0)}, metric='euclid', copy_data=False)
Also you can build a periodic grid in the same step
[6]:
gsp = GriSPy(data, periodic=periodic)
Query for neighbors within upper_radii¶
[7]:
upper_radii = 10.0
bubble_dist, bubble_ind = gsp.bubble_neighbors(
centres, distance_upper_bound=upper_radii
)
Query for neighbors in a shell within lower_radii and upper_radii¶
[8]:
upper_radii = 10.0
lower_radii = 8.0
shell_dist, shell_ind = gsp.shell_neighbors(
centres,
distance_lower_bound=lower_radii,
distance_upper_bound=upper_radii
)
Query for nth nearest neighbors¶
[9]:
n_nearest = 10
near_dist, near_ind = gsp.nearest_neighbors(centres, n=n_nearest)
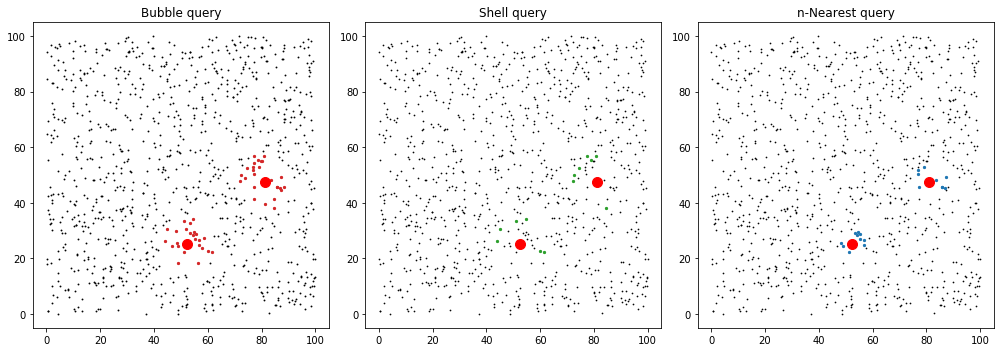
Plot results¶
[10]:
fig, axes = plt.subplots(1, 3, figsize=(14, 5))
ax = axes[0]
ax.set_title("Bubble query")
ax.scatter(data[:, 0], data[:, 1], c="k", marker=".", s=3)
for ind in bubble_ind:
ax.scatter(data[ind, 0], data[ind, 1], c="C3", marker="o", s=5)
ax.plot(centres[:,0],centres[:,1],'ro',ms=10)
ax = axes[1]
ax.set_title("Shell query")
ax.scatter(data[:, 0], data[:, 1], c="k", marker=".", s=2)
for ind in shell_ind:
ax.scatter(data[ind, 0], data[ind, 1], c="C2", marker="o", s=5)
ax.plot(centres[:,0],centres[:,1],'ro',ms=10)
ax = axes[2]
ax.set_title("n-Nearest query")
ax.scatter(data[:, 0], data[:, 1], c="k", marker=".", s=2)
for ind in near_ind:
ax.scatter(data[ind, 0], data[ind, 1], c="C0", marker="o", s=5)
ax.plot(centres[:,0],centres[:,1],'ro',ms=10)
fig.tight_layout()
Creating your curstom distance function¶
Let’s assume that we intend to compare our distances using levenshtein’s metric for similarity between text (https://en.wikipedia.org/wiki/Levenshtein_distance).
Luckly we have the excellent textdistance library that implements efficiently this distance.
We can install it with
$ pip install textdistance
and then import it with
[11]:
import textdistance
So to make these custom distance compatible with GriSPy, we must define a function that receives 3 parameters: - c0 the center to which we seek the distance. - centres the \(C\) centers to which we want to calculate the distance from a c0. - dim the dimension of each center and c0.
Finally the function must return a np.ndarray with \(C\) elements where the element \(j-nth\) corresponds to the distance between c0 and centres\(_j\).
[12]:
def levenshtein(c0, centres, dim):
# textdistance only operates over list and tuples
c0 = tuple(c0)
# creates a empty array with the required
# number of distances
distances = np.empty(len(centres))
for idx, c1 in enumerate(centres):
# textdistance only operates over list and tuples
c1 = tuple(c1)
# calculate the distance
dis = textdistance.levenshtein(c0, c1)
# store the distance
distances[idx] = dis
return distances
Then we create the grid with the custom distance, and run the code
[13]:
gsp = GriSPy(data, metric=levenshtein)
upper_radii = 10.0
lev_dist, lev_ind = gsp.bubble_neighbors(
centres, distance_upper_bound=upper_radii)
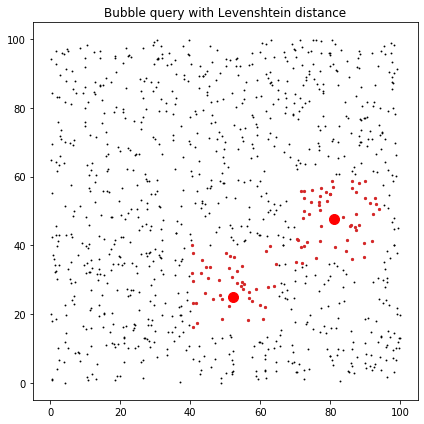
Finally we can check our bubble_neighbors result with a plot
[14]:
fig, axes = plt.subplots(figsize=(6, 6))
ax = axes
ax.set_title("Bubble query with Levenshtein distance")
ax.scatter(data[:, 0], data[:, 1], c="k", marker=".", s=3)
for ind in lev_ind:
ax.scatter(data[ind, 0], data[ind, 1], c="C3", marker="o", s=5)
ax.plot(centres[:,0],centres[:,1],'ro',ms=10)
fig.tight_layout()